Note
Click here to download the full example code
reclustering on specific leiden clusters¶
This example demonstrates who to perform a reclustering on a selected subset of leiden clusters. You will want to do this for example during the process of celltype annotation, when the clusters do not have a sufficient resolution to seperate all clusters and mixed cell populations still exist.
import besca as bc
import scanpy as sc
import pytest
# pytest.skip('Test is only for here as example and should not be executed')
# load and preprocess data (here we will start from a preprocessed dataset)
adata = bc.datasets.pbmc3k_processed()
# extract subset using the recluster function whcih is part of the reclustering (rc) toolkit
adata_subset = bc.tl.rc.recluster(
adata,
celltype=("2", "3", "4", "5", "6", "8", "9", "10", "11", "12"),
celltype_label="leiden",
resolution=1.2,
)
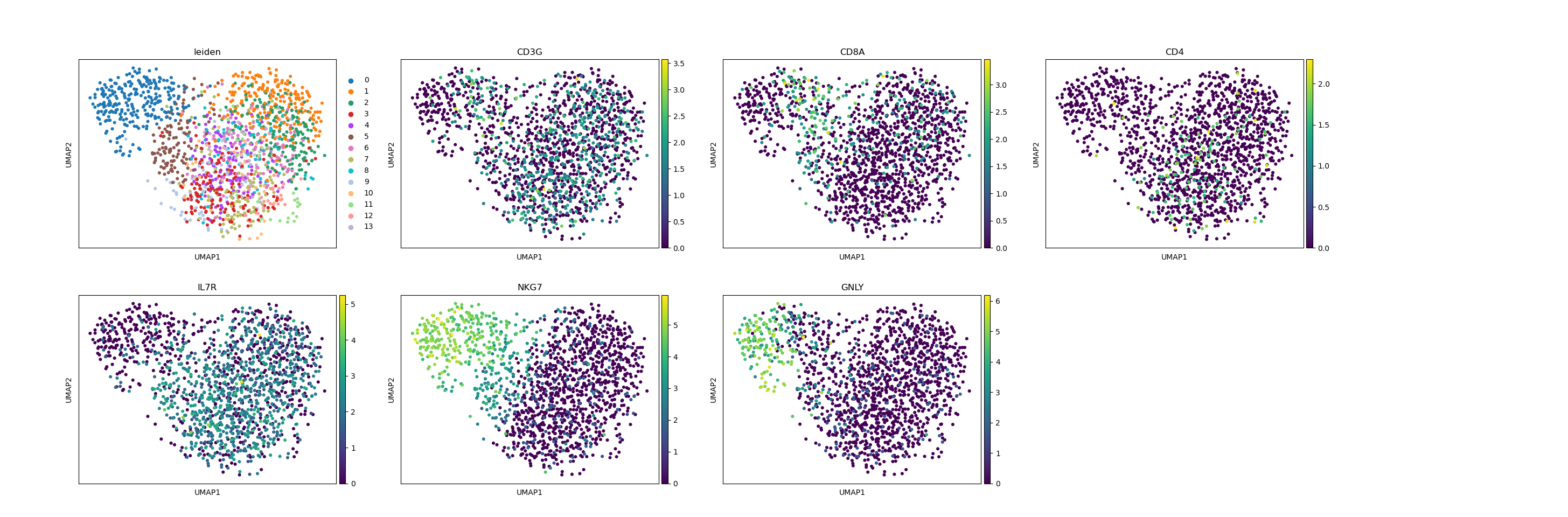
# visualize the new clusters
sc.pl.umap(
adata_subset, color=["leiden", "CD3G", "CD8A", "CD4", "IL7R", "NKG7", "GNLY"]
)
# append new celltype labels to the subclusters.
# This is an approximative hand annotation that should be dealt into more widths.
labels = [
"NK cell", # 0
"CD4 T-cell", # 1
"CD8 T-cell", # 2
"CD4 T-cell", # 3
"CD8 T-cell", # 4
"CD8 T-cell", # 5
"CD4 T-cell", # 6
"CD4 T-cell", # 7
"CD4 T-cell", # 8
"CD4 T-cell", # 9
"CD4 T-cell", # 10
"CD4 T-cell", # 11
"CD4 T-cell", # 12
"CD4 T-cell", # 13
"CD4 T-cell", # 14
"CD4 T-cell", # 15
"CD4 T-cell", # 16
"CD4 T-cell", # 17
"CD4 T-cell", # 18
"CD4 T-cell", # 19
] # 10
new_labels = [
labels[i]
for i in range(len(adata_subset.obs.get("leiden").value_counts().index.tolist()))
]
# merge the labels back into the original adata object
# note this will overwrite what ever was saved in adata.obs.celltype;
# Here is was not assigned yet.
bc.tl.rc.annotate_new_cellnames(
adata, adata_subset, names=new_labels, new_label="celltype"
)
Total running time of the script: ( 0 minutes 3.241 seconds)