Automated Annotation Workflow¶
This workflow uses the auto_annot tools from besca to newly annotate a scRNAseq dataset based on one or more preannotated datasets. Ideally, these datasets come from a similar tissue and condition. If multiple training datasets are used, the performance is dependent on all dataset being annotated to the same resolution with broadly similar cell types.
We use supervised machine learning methods to annotate each individual cell utilizing methods like support vector machines (SVM) or logistic regression.
First, the traning dataset(s) and the testing dataset are loaded from h5ad files or made available as adata objects. Next, the training and testing datasets are corrected using scanorama, and the training datasets are then merged into one anndata object. Then, the classifier is trained utilizing the merged training data. Finally, the classifier is applied to the testing dataset to predict the cell types. If the testing dataset is already annotated (to test the algorithm), a report including confusion matrices can be generated.
[1]:
import besca as bc
Parameter specification¶
Give your analysis a name.
[2]:
analysis_name = (
"auto_annot_tutorial" # The analysis name will be used to name the output files
)
Specify column name of celltype annotation you want to train on.
[3]:
celltype = "dblabel" # This needs to be a column in the .obs of the training datasets (and test dataset if you want to generate a report)
Choose a method:
linear: Support Vector Machine with Linear Kernel
sgd: Support Vector Machine with Linear Kernel using Stochastic Gradient Descent
rbf: Support Vector Machine with radial basis function kernel. Very time intensive, use only on small datasets.
logistic_regression: Standard logistic classifier iwth multinomial loss.
logistic_regression_ovr: Logistic Regression with one versus rest classification.
logistic_regression_elastic: Logistic Regression with elastic loss, cross validates among multiple l1 ratios.
[4]:
method = "logistic_regression"
Specify merge method. Needs to be either scanorama or naive.
[5]:
merge = "scanorama" # We recommend to use scanorama here
Decide if you want to use the raw format or highly variable genes. Raw increases computational time and does not necessarily improve predictions.
[6]:
use_raw = False # We recommend to use False here
You can choose to only consider a subset of genes from a signature set or use all genes.
[7]:
genes_to_use = "all" # We suggest to use all here, but the runtime is strongly improved if you select an appropriate gene set
Data loading¶
Read in all training sets and the testing set.¶
We will use a publicly available PBMC dataset, including ~3000 cells, as testing dataset. For the training dataset we use another PBMC dataset that is delivered with besca. The training datasets used are from:
Granja JM, Klemm S, McGinnis LM, et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat Biotechnol. 2019;37(12):1458-1465. doi:10.1038/s41587-019-0332-7
and
Kotliarov Y, Sparks R, Martins AJ, et al. Broad immune activation underlies shared set point signatures for vaccine responsiveness in healthy individuals and disease activity in patients with lupus. Nat Med. 2020;26(4):618-629. doi:10.1038/s41591-020-0769-8
A detailed analysis of the annotation performed below can be found in figure 3 of the publication on besca.
[8]:
adata_test = (
bc.datasets.pbmc3k_processed()
) # Dataset to be annotated (it's already annotated in this case for testing)
adata_test_orig = (
adata_test.copy()
) # Make a copy of the testing data, which will be annotated in .obs, but not further modified
adata_train_list = [
bc.datasets.Granja2019_processed(),
bc.datasets.Kotliarov2020_processed(),
] # List of annotated training datasets
You can also load your own dataset from the file system. Specify folders where .h5ad files are found and their names. The datasets that are already annotated and should be used for training. If you only use one dataset please use list of one. These load functions are useful in particular when the datasets have been processed with the besca standard pipeline. Alternatively, datasets can be loaded in any fashion, if more suitable.
[9]:
# test_dataset_path = '/path/to/test/dataset/folder'
# test_dataset = 'testdataset.h5ad'
#
# train_dataset_paths = ['/path/to/train/dataset/folder1', '/path/to/train/dataset/folder2']
# train_datasets = ['traindataset1.h5ad', 'traindataset2.h5ad']
[10]:
# adata_train_list, adata_test, adata_test_orig = bc.tl.auto_annot.read_data(train_paths = train_dataset_paths, train_datasets= train_datasets, test_path= test_dataset_path, test_dataset= test_dataset, use_raw = use_raw)
One training dataset’s label column is not named “dblabel”. We need to create a column so that all datasets used contain a column that are named according to the previously specified variable celltype and that contain the labels.
[11]:
adata_train_list[1].obs["dblabel"] = adata_train_list[1].obs["celltype3"]
In this case the testing dataset is already annotated to demonstrate the methodology. All datasets adhere to the same naming convention.
[12]:
adata_test.obs.dblabel.unique()
[12]:
['naive thymus-derived CD8-positive, alpha-beta..., 'naive B cell', 'central memory CD4-positive, alpha-beta T cell', 'classical monocyte', 'IL7R-max CD8-positive, alpha-beta cytotoxic T..., 'non-classical monocyte', 'naive thymus-derived CD4-positive, alpha-beta..., 'CD8-positive, alpha-beta cytotoxic T cell', 'cytotoxic CD56-dim natural killer cell', 'CD1c-positive myeloid dendritic cell']
Categories (10, object): ['CD1c-positive myeloid dendritic cell', 'CD8-positive, alpha-beta cytotoxic T cell', 'IL7R-max CD8-positive, alpha-beta cytotoxic T..., 'central memory CD4-positive, alpha-beta T cell', ..., 'naive B cell', 'naive thymus-derived CD4-positive, alpha-beta..., 'naive thymus-derived CD8-positive, alpha-beta..., 'non-classical monocyte']
[13]:
adata_train_list[0].obs.dblabel.unique()
[13]:
['naive thymus-derived CD4-positive, alpha-beta..., 'classical monocyte', 'naive B cell', 'lymphocyte of B lineage', 'naive thymus-derived CD8-positive, alpha-beta..., ..., 'IL7R-max CD8-positive, alpha-beta cytotoxic T..., 'hematopoietic multipotent progenitor cell', 'myeloid leukocyte', 'basophil', 'plasma cell']
Length: 25
Categories (25, object): ['naive thymus-derived CD4-positive, alpha-beta..., 'naive thymus-derived CD8-positive, alpha-beta..., 'naive B cell', 'IL7R-max CD8-positive, alpha-beta cytotoxic T..., ..., 'common lymphoid progenitor', 'basophil', 'plasma cell', 'erythrocyte']
[14]:
adata_train_list[1].obs.dblabel.unique()
[14]:
['cytotoxic CD56-dim natural killer cell', 'naive thymus-derived CD8-positive, alpha-beta..., 'naive thymus-derived CD4-positive, alpha-beta..., 'classical monocyte', 'CD8-positive, alpha-beta cytotoxic T cell', ..., 'regulatory T cell', 'CD1c-positive myeloid dendritic cell', 'plasmacytoid dendritic cell', 'erythrocyte', 'plasma cell']
Length: 14
Categories (14, object): ['cytotoxic CD56-dim natural killer cell', 'naive B cell', 'IL7R-max CD8-positive, alpha-beta cytotoxic T..., 'memory B cell', ..., 'plasmacytoid dendritic cell', 'erythrocyte', 'plasma cell', 'naive thymus-derived CD8-positive, alpha-beta...]
Correct datasets (e.g. using scanorama) and merge training datasets¶
This function merges training datasets, removes unwanted genes, and if scanorama is used corrects for datasets.
[15]:
adata_train, adata_test_corrected = bc.tl.auto_annot.merge_data(
adata_train_list, adata_test, genes_to_use=genes_to_use, merge=merge
)
merging with scanorama
using scanorama rn
Found 207 genes among all datasets
[[0. 0.69861833 0.47963259]
[0. 0. 0.98881789]
[0. 0. 0. ]]
Processing datasets (1, 2)
Processing datasets (0, 1)
Processing datasets (0, 2)
integrating training set
Train the classifier¶
The returned scaler is fitted on the training dataset (to zero mean and scaled to unit variance). The scaling will then be applied to the counts in the testing dataset and then the classifier is applied to the scaled testing dataset (see next step, adata_predict()). This function will run multiple jobs in parallel if if logistic regression was specified as method.
[16]:
classifier, scaler = bc.tl.auto_annot.fit(adata_train, method, celltype, njobs=14)
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 2.12039D+05 |proj g|= 2.86381D+04
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 2.11969D+05 |proj g|= 2.70303D+04
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 2.11946D+05 |proj g|= 2.74976D+04
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 2.12051D+05 |proj g|= 2.70821D+04
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 2.11957D+05 |proj g|= 2.71735D+04
At iterate 50 f= 1.04971D+05 |proj g|= 2.73525D+01
At iterate 50 f= 1.03637D+05 |proj g|= 8.64883D+00
At iterate 50 f= 1.03011D+05 |proj g|= 8.89155D+00
At iterate 50 f= 1.03004D+05 |proj g|= 5.90025D+00
At iterate 50 f= 1.04246D+05 |proj g|= 7.64672D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 70 73 1 0 0 1.162D+00 1.030D+05
F = 103010.68989000401
CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 7.74054D+04 |proj g|= 2.54100D+03
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 72 76 1 0 0 1.197D+00 1.050D+05
F = 104971.11070892689
CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 7.96649D+04 |proj g|= 2.47790D+03
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 74 78 1 0 0 6.220D-01 1.036D+05
F = 103636.52996705240
CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 7.83556D+04 |proj g|= 2.44585D+03
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 77 82 1 0 0 6.687D-01 1.030D+05
F = 103003.76722693436
CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 7.76244D+04 |proj g|= 2.55120D+03
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 80 85 1 0 0 8.110D-01 1.042D+05
F = 104245.97290945785
CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 7.90204D+04 |proj g|= 2.50165D+03
At iterate 50 f= 5.93638D+04 |proj g|= 2.00635D+01
At iterate 50 f= 6.15150D+04 |proj g|= 3.03599D+01
At iterate 50 f= 6.04466D+04 |proj g|= 1.57444D+01
At iterate 50 f= 5.98289D+04 |proj g|= 2.91009D+01
At iterate 50 f= 6.11011D+04 |proj g|= 2.25134D+01
At iterate 100 f= 5.93575D+04 |proj g|= 4.64568D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 104 1 0 0 4.646D+00 5.936D+04
F = 59357.500019558975
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 4.73539D+04 |proj g|= 5.57261D+02
At iterate 100 f= 6.15099D+04 |proj g|= 1.67961D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 1.680D+00 6.151D+04
F = 61509.900886338968
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 4.94544D+04 |proj g|= 6.00582D+02
At iterate 100 f= 6.04395D+04 |proj g|= 1.89094D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 1.891D+00 6.044D+04
F = 60439.492975334550
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 4.85205D+04 |proj g|= 5.86978D+02
At iterate 100 f= 5.98230D+04 |proj g|= 1.48425D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 1.484D+00 5.982D+04
F = 59822.961628412340
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 4.79571D+04 |proj g|= 5.79302D+02
At iterate 100 f= 6.10936D+04 |proj g|= 2.97156D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 108 1 0 0 2.972D+00 6.109D+04
F = 61093.551063188512
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 4.91858D+04 |proj g|= 5.88940D+02
At iterate 50 f= 4.11176D+04 |proj g|= 2.70626D+01
At iterate 50 f= 4.31390D+04 |proj g|= 3.15829D+01
At iterate 50 f= 4.23066D+04 |proj g|= 3.96669D+01
At iterate 50 f= 4.17778D+04 |proj g|= 2.30889D+01
At iterate 50 f= 4.30381D+04 |proj g|= 2.38411D+01
At iterate 100 f= 4.10888D+04 |proj g|= 7.57278D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 104 1 0 0 7.573D+00 4.109D+04
F = 41088.792765115402
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.64715D+04 |proj g|= 1.14671D+02
At iterate 100 f= 4.22764D+04 |proj g|= 1.94224D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 1.942D+01 4.228D+04
F = 42276.428932985196
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 4.31078D+04 |proj g|= 2.42772D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 108 1 0 0 2.428D+01 4.311D+04
F = 43107.796510953172
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.76858D+04 |proj g|= 1.23967D+02
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.84692D+04 |proj g|= 1.18329D+02
At iterate 100 f= 4.17517D+04 |proj g|= 4.54061D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 105 1 0 0 4.541D+01 4.175D+04
F = 41751.728515726456
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.71857D+04 |proj g|= 1.24753D+02
At iterate 100 f= 4.30019D+04 |proj g|= 1.31606D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 103 1 0 0 1.316D+01 4.300D+04
F = 43001.948646084813
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.84556D+04 |proj g|= 1.19735D+02
At iterate 50 f= 3.43179D+04 |proj g|= 1.61990D+01
At iterate 50 f= 3.55724D+04 |proj g|= 5.30592D+01
At iterate 50 f= 3.63519D+04 |proj g|= 2.47038D+01
At iterate 50 f= 3.50436D+04 |proj g|= 3.81051D+01
At iterate 50 f= 3.63920D+04 |proj g|= 5.06944D+01
At iterate 100 f= 3.42672D+04 |proj g|= 1.54011D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 105 1 0 0 1.540D+01 3.427D+04
F = 34267.225020703758
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.26307D+04 |proj g|= 2.29930D+01
At iterate 100 f= 3.62979D+04 |proj g|= 6.97876D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 6.979D+01 3.630D+04
F = 36297.920086924394
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 3.55243D+04 |proj g|= 2.52911D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 2.529D+01 3.552D+04
F = 35524.276263567997
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.46728D+04 |proj g|= 7.72150D+01
At iterate 100 f= 3.49969D+04 |proj g|= 9.13133D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 104 1 0 0 9.131D+00 3.500D+04
F = 34996.943723823075
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.39185D+04 |proj g|= 3.31235D+01
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.33719D+04 |proj g|= 2.34712D+01
At iterate 100 f= 3.63334D+04 |proj g|= 1.43529D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 1.435D+01 3.633D+04
F = 36333.443243589172
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.47128D+04 |proj g|= 2.46425D+01
At iterate 50 f= 3.20623D+04 |proj g|= 5.45743D+01
At iterate 50 f= 3.41204D+04 |proj g|= 3.54927D+01
At iterate 50 f= 3.33592D+04 |proj g|= 3.47271D+01
At iterate 50 f= 3.27833D+04 |proj g|= 5.03492D+01
At iterate 50 f= 3.41880D+04 |proj g|= 8.07172D+01
At iterate 100 f= 3.19593D+04 |proj g|= 5.11212D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 110 1 0 0 5.112D+01 3.196D+04
F = 31959.310456641466
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.14746D+04 |proj g|= 5.22624D+01
At iterate 100 f= 3.40062D+04 |proj g|= 1.23469D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 1.235D+01 3.401D+04
F = 34006.179329086517
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.35020D+04 |proj g|= 1.28782D+01
At iterate 100 f= 3.32580D+04 |proj g|= 2.06409D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 2.064D+01 3.326D+04
F = 33258.003907179125
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 3.26660D+04 |proj g|= 1.49031D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 108 1 0 0 1.490D+01 3.267D+04
F = 32666.015817912816
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.27945D+04 |proj g|= 2.22048D+01
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.21525D+04 |proj g|= 1.54769D+01
At iterate 100 f= 3.40856D+04 |proj g|= 2.61951D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 108 1 0 0 2.620D+01 3.409D+04
F = 34085.608687699583
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.36314D+04 |proj g|= 2.70166D+01
At iterate 50 f= 3.13759D+04 |proj g|= 5.13549D+01
At iterate 50 f= 3.26863D+04 |proj g|= 4.69096D+01
At iterate 50 f= 3.34000D+04 |proj g|= 2.70486D+01
At iterate 50 f= 3.20391D+04 |proj g|= 1.90035D+01
At iterate 50 f= 3.35226D+04 |proj g|= 9.19913D+01
At iterate 100 f= 3.12623D+04 |proj g|= 2.61464D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 109 1 0 0 2.615D+01 3.126D+04
F = 31262.281830163720
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.11541D+04 |proj g|= 2.60008D+01
At iterate 100 f= 3.25599D+04 |proj g|= 4.23087D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 4.231D+01 3.256D+04
F = 32559.924042316710
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.24459D+04 |proj g|= 4.21871D+01
At iterate 100 f= 3.32669D+04 |proj g|= 1.81318D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 1.813D+01 3.327D+04
F = 33266.879532518455
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.31391D+04 |proj g|= 1.78562D+01
At iterate 100 f= 3.18701D+04 |proj g|= 2.01354D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 108 1 0 0 2.014D+01 3.187D+04
F = 31870.144245662003
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.17202D+04 |proj g|= 2.02726D+01
At iterate 100 f= 3.33875D+04 |proj g|= 3.87361D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 105 1 0 0 3.874D+01 3.339D+04
F = 33387.501525823609
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.32710D+04 |proj g|= 3.89076D+01
At iterate 50 f= 3.24120D+04 |proj g|= 1.68045D+01
At iterate 50 f= 3.11244D+04 |proj g|= 2.11394D+01
At iterate 50 f= 3.31137D+04 |proj g|= 1.36011D+01
At iterate 50 f= 3.16790D+04 |proj g|= 1.49819D+01
At iterate 50 f= 3.32350D+04 |proj g|= 2.77797D+01
At iterate 100 f= 3.10458D+04 |proj g|= 5.27721D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 5.277D+01 3.105D+04
F = 31045.808935526016
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 3.23504D+04 |proj g|= 1.68157D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 1.682D+01 3.235D+04
F = 32350.360595808739
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.10268D+04 |proj g|= 5.27721D+01
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.23327D+04 |proj g|= 1.68403D+01
At iterate 100 f= 3.16356D+04 |proj g|= 2.32266D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 2.323D+01 3.164D+04
F = 31635.616571226965
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 3.30666D+04 |proj g|= 2.88477D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 108 1 0 0 2.885D+01 3.307D+04
F = 33066.577323674304
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.16146D+04 |proj g|= 2.32253D+01
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.30481D+04 |proj g|= 2.88754D+01
At iterate 100 f= 3.31910D+04 |proj g|= 1.95868D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 1.959D+01 3.319D+04
F = 33190.978410728567
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.31747D+04 |proj g|= 1.95730D+01
At iterate 50 f= 3.23028D+04 |proj g|= 1.67149D+01
At iterate 50 f= 3.10050D+04 |proj g|= 6.82002D+00
At iterate 50 f= 3.15900D+04 |proj g|= 1.27052D+01
At iterate 50 f= 3.30243D+04 |proj g|= 8.12354D+00
At iterate 50 f= 3.31546D+04 |proj g|= 2.98043D+01
At iterate 100 f= 3.22851D+04 |proj g|= 2.58396D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 2.584D+01 3.229D+04
F = 32285.126911492029
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.22824D+04 |proj g|= 2.58431D+01
At iterate 100 f= 3.09928D+04 |proj g|= 7.38270D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 109 1 0 0 7.383D+00 3.099D+04
F = 30992.812399692764
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 3.15678D+04 |proj g|= 1.11096D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 105 1 0 0 1.111D+01 3.157D+04
F = 31567.796015751748
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.15645D+04 |proj g|= 1.11109D+01
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.09901D+04 |proj g|= 7.38042D+00
At iterate 100 f= 3.30095D+04 |proj g|= 6.40468D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 6.405D+00 3.301D+04
F = 33009.483212087900
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.30067D+04 |proj g|= 6.40487D+00
At iterate 100 f= 3.31285D+04 |proj g|= 1.18847D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 1.188D+01 3.313D+04
F = 33128.459170482820
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.31258D+04 |proj g|= 1.18797D+01
At iterate 50 f= 3.22709D+04 |proj g|= 2.02002D+01
At iterate 50 f= 3.09805D+04 |proj g|= 5.53911D+00
At iterate 50 f= 3.15491D+04 |proj g|= 8.27267D+00
At iterate 50 f= 3.29966D+04 |proj g|= 1.00619D+01
At iterate 50 f= 3.31086D+04 |proj g|= 6.37572D+00
At iterate 100 f= 3.22532D+04 |proj g|= 1.12258D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 1.123D+01 3.225D+04
F = 32253.171229828164
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.22528D+04 |proj g|= 1.12254D+01
At iterate 100 f= 3.15356D+04 |proj g|= 1.73320D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 105 1 0 0 1.733D+01 3.154D+04
F = 31535.570385863415
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 3.09676D+04 |proj g|= 1.96023D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 1.960D+01 3.097D+04
F = 30967.570517159849
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.15351D+04 |proj g|= 1.73315D+01
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.09672D+04 |proj g|= 1.96027D+01
At iterate 100 f= 3.29838D+04 |proj g|= 4.59068D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 4.591D+00 3.298D+04
F = 32983.762119559331
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.29834D+04 |proj g|= 4.59065D+00
At iterate 100 f= 3.30958D+04 |proj g|= 4.64356D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 4.644D+01 3.310D+04
F = 33095.848676494395
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
RUNNING THE L-BFGS-B CODE
* * *
Machine precision = 2.220D-16
N = 5200 M = 10
At X0 0 variables are exactly at the bounds
At iterate 0 f= 3.30955D+04 |proj g|= 4.64353D+01
At iterate 50 f= 3.22430D+04 |proj g|= 6.39587D+00
At iterate 50 f= 3.15266D+04 |proj g|= 1.91596D+01
At iterate 50 f= 3.09581D+04 |proj g|= 4.43404D+00
At iterate 50 f= 3.29741D+04 |proj g|= 1.85344D+01
At iterate 50 f= 3.30852D+04 |proj g|= 8.04640D+00
At iterate 100 f= 3.22340D+04 |proj g|= 7.42192D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 7.422D+00 3.223D+04
F = 32234.027868082881
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 3.15146D+04 |proj g|= 6.80253D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 107 1 0 0 6.803D+00 3.151D+04
F = 31514.581161989288
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 3.09528D+04 |proj g|= 6.39301D+00
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 110 1 0 0 6.393D+00 3.095D+04
F = 30952.786412086960
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 3.29657D+04 |proj g|= 1.48928D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 108 1 0 0 1.489D+01 3.297D+04
F = 32965.669129274691
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
At iterate 100 f= 3.30737D+04 |proj g|= 1.99126D+01
* * *
Tit = total number of iterations
Tnf = total number of function evaluations
Tnint = total number of segments explored during Cauchy searches
Skip = number of BFGS updates skipped
Nact = number of active bounds at final generalized Cauchy point
Projg = norm of the final projected gradient
F = final function value
* * *
N Tit Tnf Tnint Skip Nact Projg F
5200 100 106 1 0 0 1.991D+01 3.307D+04
F = 33073.655496377971
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT
Prediction¶
Use fitted model to predict celltypes in adata_pred (adata_test_corrected in our case). In the case of logistic regression, the threshold specifies the probability that needs to be reached to annotate a cell type or will be annotated as “unknown” if not reached. The threshold should be set to 0 or left out for SVM.
[17]:
adata_predicted = bc.tl.auto_annot.adata_predict(
classifier=classifier,
scaler=scaler,
adata_pred=adata_test_corrected,
adata_orig=adata_test_orig,
threshold=0.1,
)
The prediction was added in a new column called ‘auto_annot’.
If in addition to the most likely class you would like to have all class probabilities returned use the following function. (This is only a sensible choice if using logistic regression.)
[18]:
adata_predicted = bc.tl.auto_annot.adata_pred_prob(
classifier=classifier,
scaler=scaler,
adata_pred=adata_test_corrected,
adata_orig=adata_test_orig,
threshold=0.1,
)
Output¶
The adata object that includes the predicted cell type annotation can be written out as h5ad file.
[19]:
adata_predicted.write("/tmp/adata_predicted.h5ad")
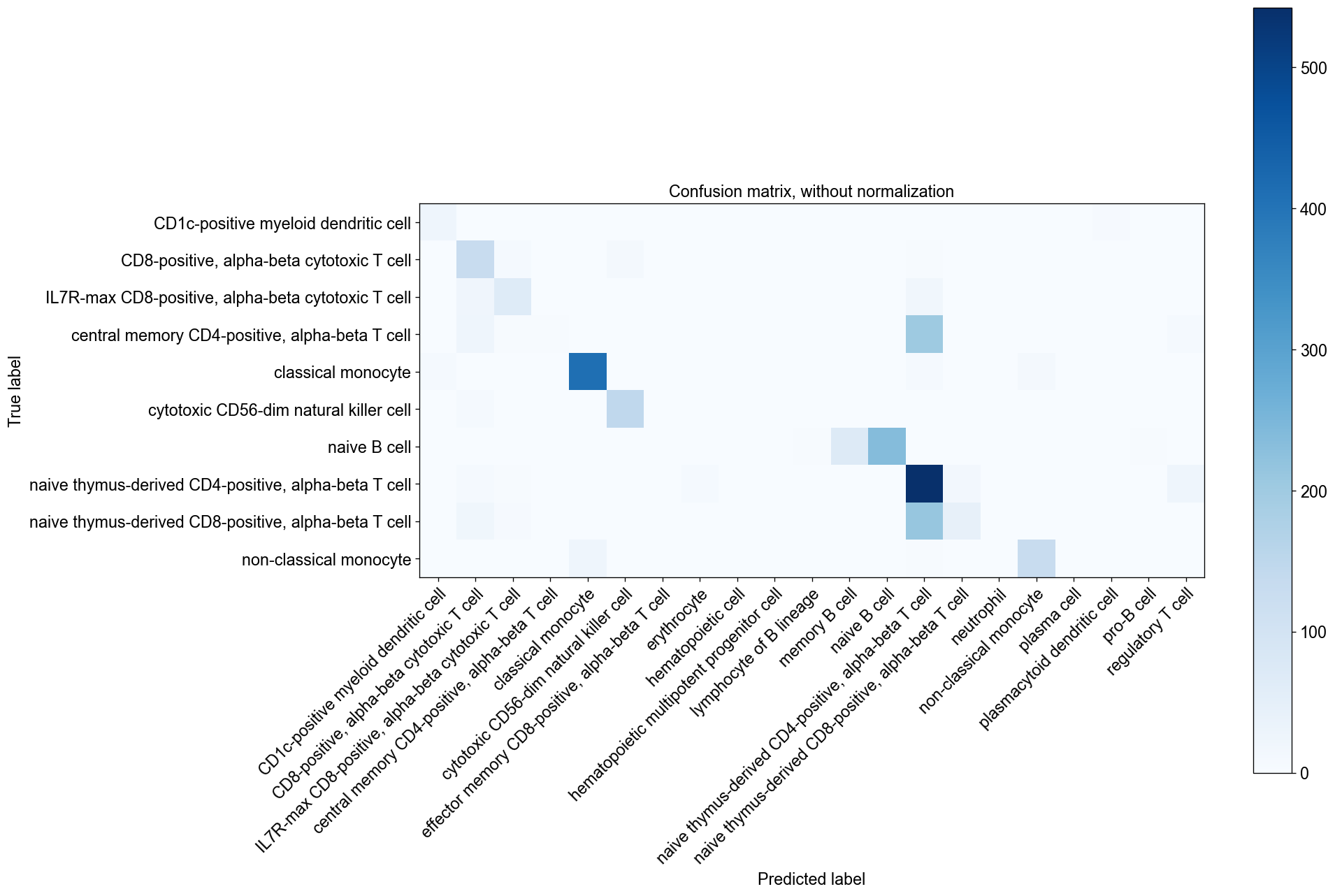
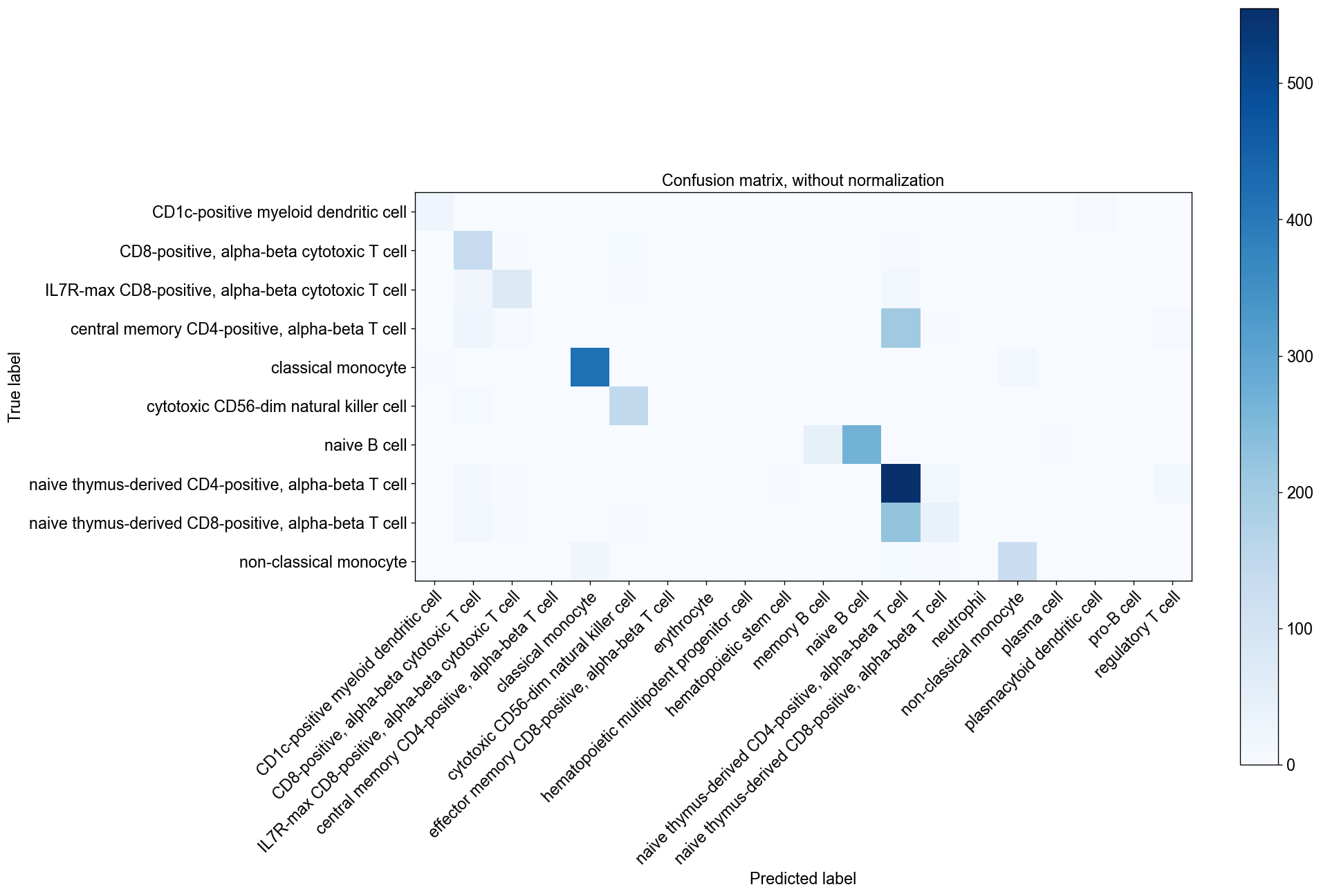
If the testing dataset included already a cell type annotation, a report can be generated and written, which includes metrics, confusion matrices and comparative umap plots. The report creates standardised overview figures, for a more detailed control of figure layout other besca and scanpy figure functions should be used.
[20]:
%matplotlib inline
bc.tl.auto_annot.report(adata_predicted, celltype, method, analysis_name, False, merge, use_raw, genes_to_use, clustering = 'leiden')
besca.tl.auto_annot.report(...) is deprecated( besca > 2.3); please use besca.tl.report(...)
WARNING: saving figure to file figures/umap.ondata_auto_annot_tutorial.png
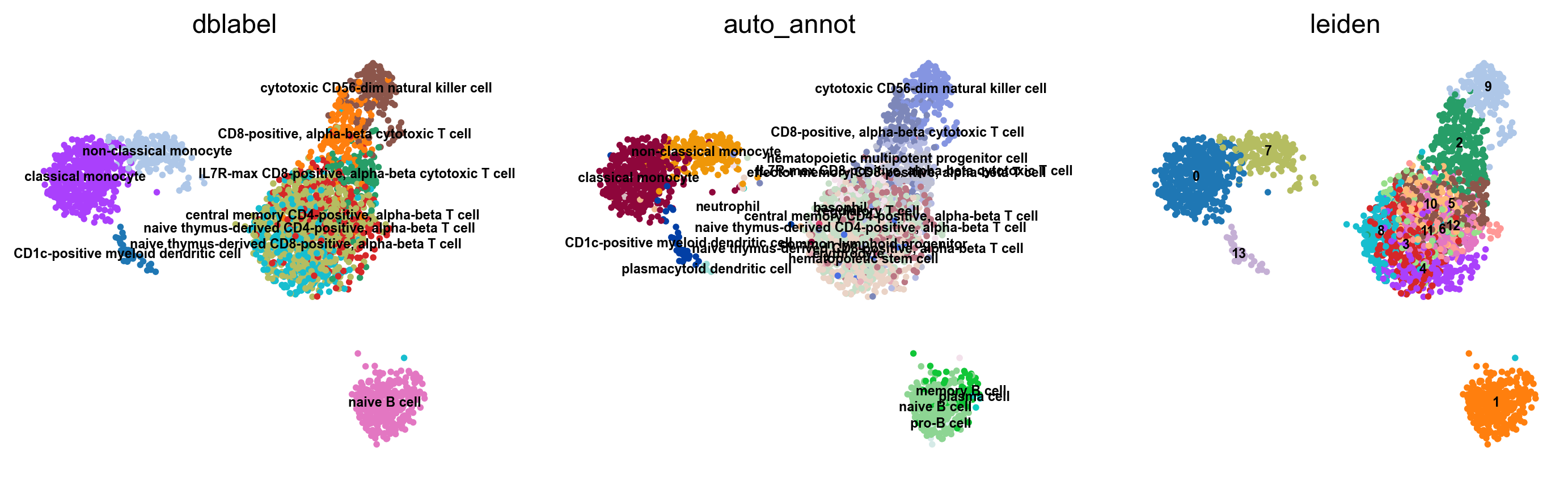
WARNING: saving figure to file figures/umap.auto_annot_tutorial_dblabel.png
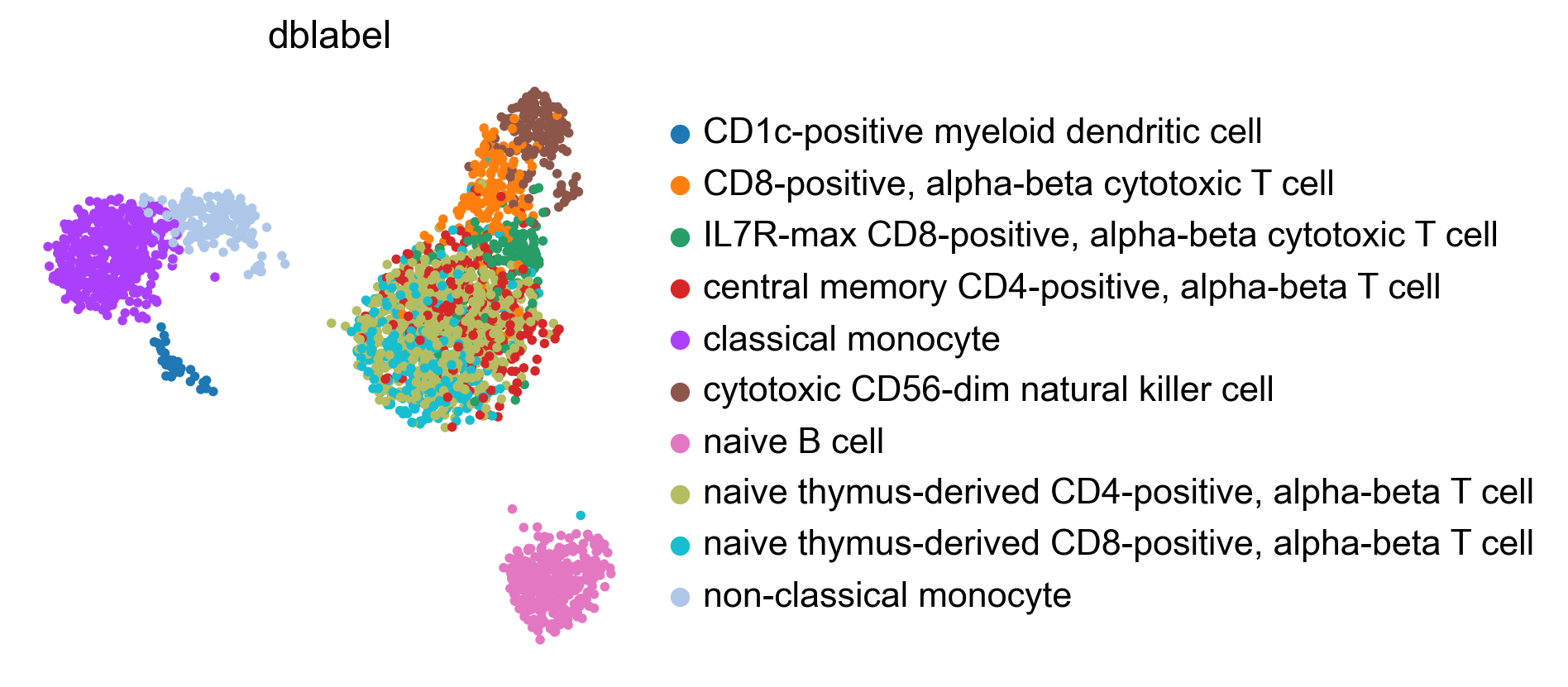
WARNING: saving figure to file figures/umap.auto_annot_tutorial_auto_annot.png
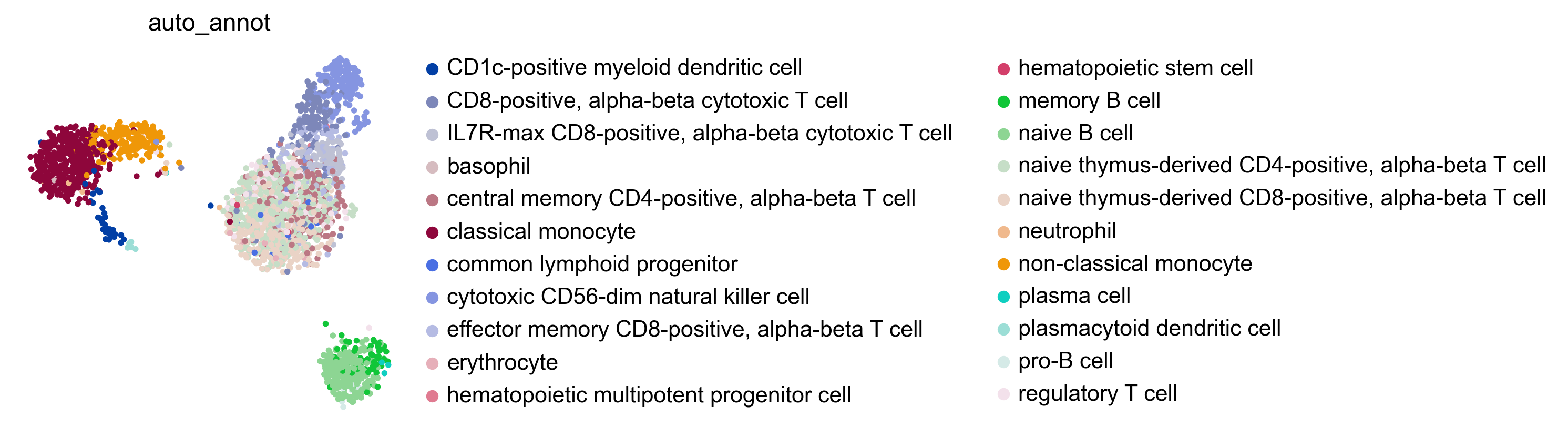
WARNING: saving figure to file figures/umap.auto_annot_tutorial_leiden.png
Data type cannot be displayed: application/vnd.plotly.v1+json


If we change our threshold, we can vary the amount of cells labelled as unknown, depending on the confidence required from the predictions. Let’s try 0.7
[21]:
adata_predicted = bc.tl.auto_annot.adata_predict(
classifier=classifier,
scaler=scaler,
adata_pred=adata_test_corrected,
adata_orig=adata_test_orig,
threshold=0.7,
)
We observe that many more cells, where the assignment is not unambiguous are now labelled as unknown.
[22]:
%matplotlib inline
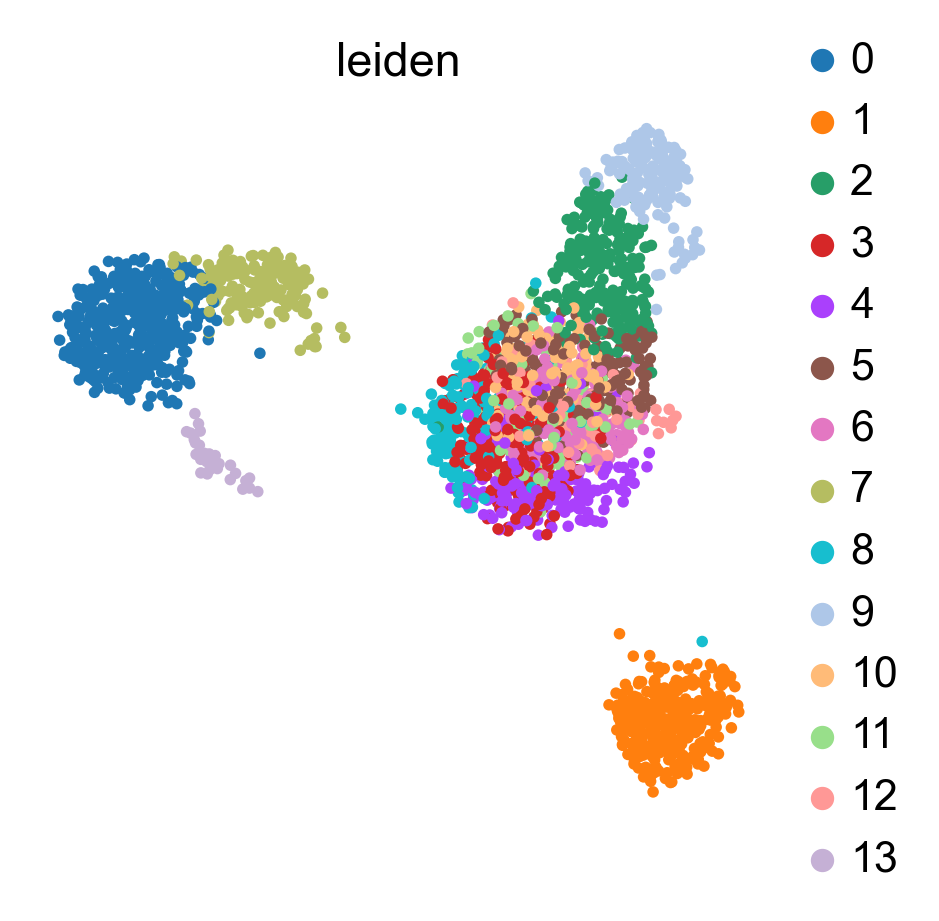
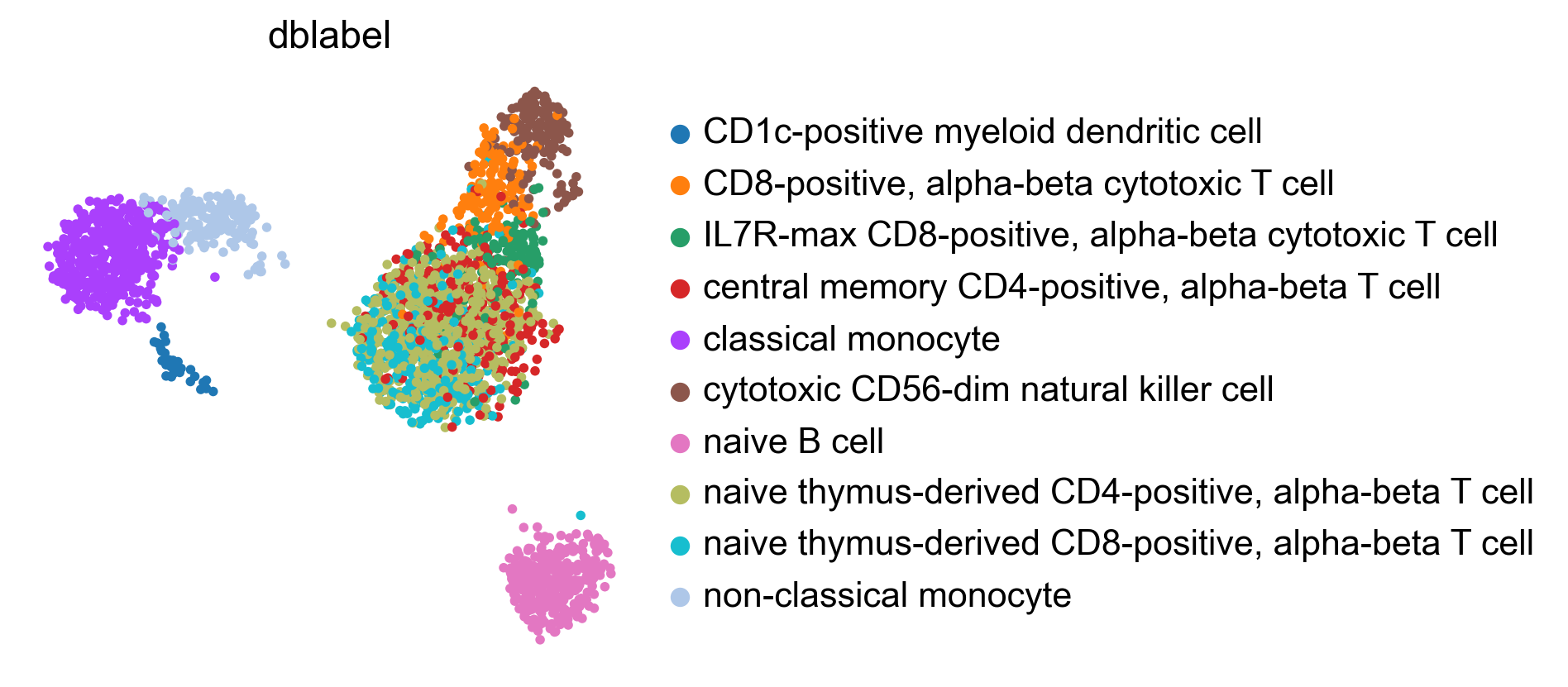
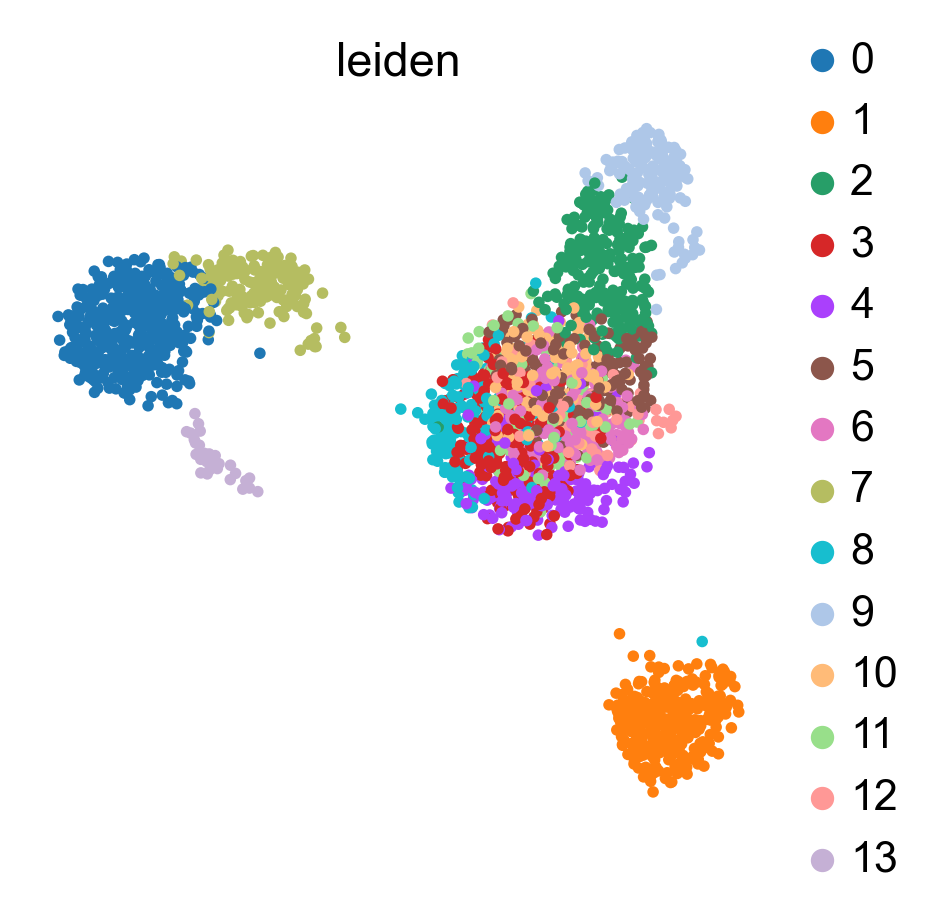
bc.tl.auto_annot.report(adata_predicted, celltype, method, analysis_name, False, merge, use_raw, genes_to_use, clustering = 'leiden')
besca.tl.auto_annot.report(...) is deprecated( besca > 2.3); please use besca.tl.report(...)
WARNING: saving figure to file figures/umap.ondata_auto_annot_tutorial.png

WARNING: saving figure to file figures/umap.auto_annot_tutorial_dblabel.png
WARNING: saving figure to file figures/umap.auto_annot_tutorial_auto_annot.png
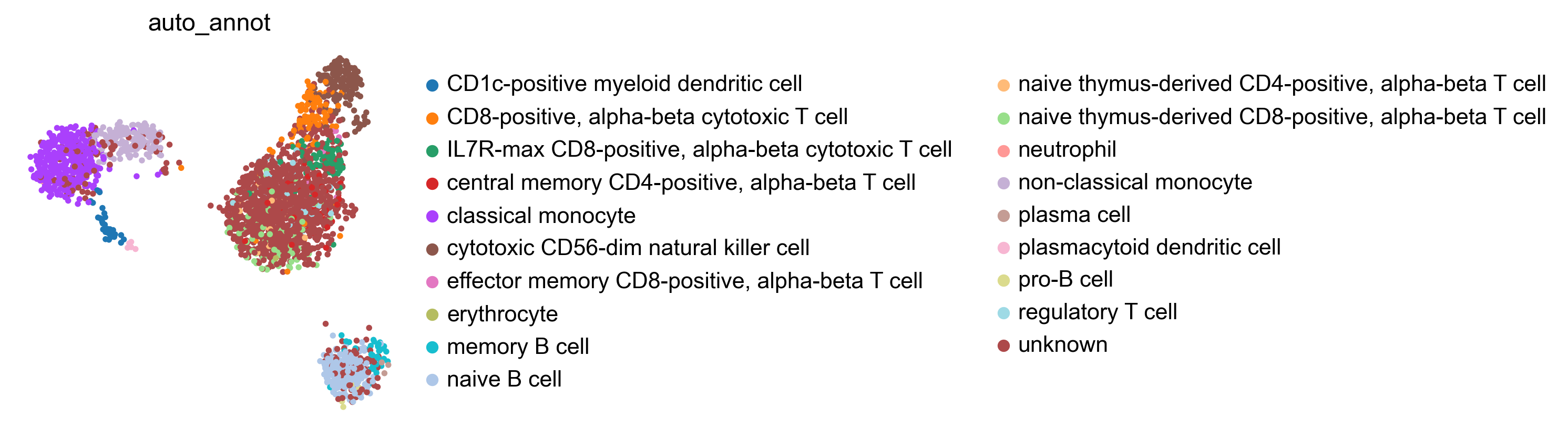
WARNING: saving figure to file figures/umap.auto_annot_tutorial_leiden.png
Data type cannot be displayed: application/vnd.plotly.v1+json


Let’s try another classifier (SVM)¶
Especially when thresholds are not used, SVMs can be tried as alternative classifiers.
[23]:
analysis_name = "auto_annot_tutorial_svm"
celltype = "dblabel" # column name of celltype of interest
method = "linear" # rbf or linear or sgd, rbf extremely slow cannot recommend, logistic_regression recommended, as you can get probabilities, random_forest is a fast but not very powerful option (in current implementation)
merge = "scanorama"
use_raw = False
genes_to_use = "all"
[24]:
adata_test = adata_test_orig.copy()
[25]:
adata_train, adata_test_corrected = bc.tl.auto_annot.merge_data(
adata_train_list, adata_test, genes_to_use=genes_to_use, merge=merge
)
merging with scanorama
using scanorama rn
Found 207 genes among all datasets
[[0. 0.7 0.48]
[0. 0. 0.99]
[0. 0. 0. ]]
Processing datasets (1, 2)
Processing datasets (0, 1)
Processing datasets (0, 2)
integrating training set
[26]:
classifier, scaler = bc.tl.auto_annot.fit(
adata_train=adata_train, method=method, celltype=celltype, njobs=14
)
[27]:
adata_predicted = bc.tl.auto_annot.adata_predict(
classifier=classifier,
scaler=scaler,
adata_pred=adata_test_corrected,
adata_orig=adata_test_orig,
)
[28]:
adata_predicted.write("/tmp/adata_predicted_svm.h5ad")
[29]:
%matplotlib inline
bc.tl.auto_annot.report(adata_predicted, celltype, method, analysis_name, False,
merge, use_raw, genes_to_use, clustering = 'leiden')
besca.tl.auto_annot.report(...) is deprecated( besca > 2.3); please use besca.tl.report(...)
WARNING: saving figure to file figures/umap.ondata_auto_annot_tutorial_svm.png
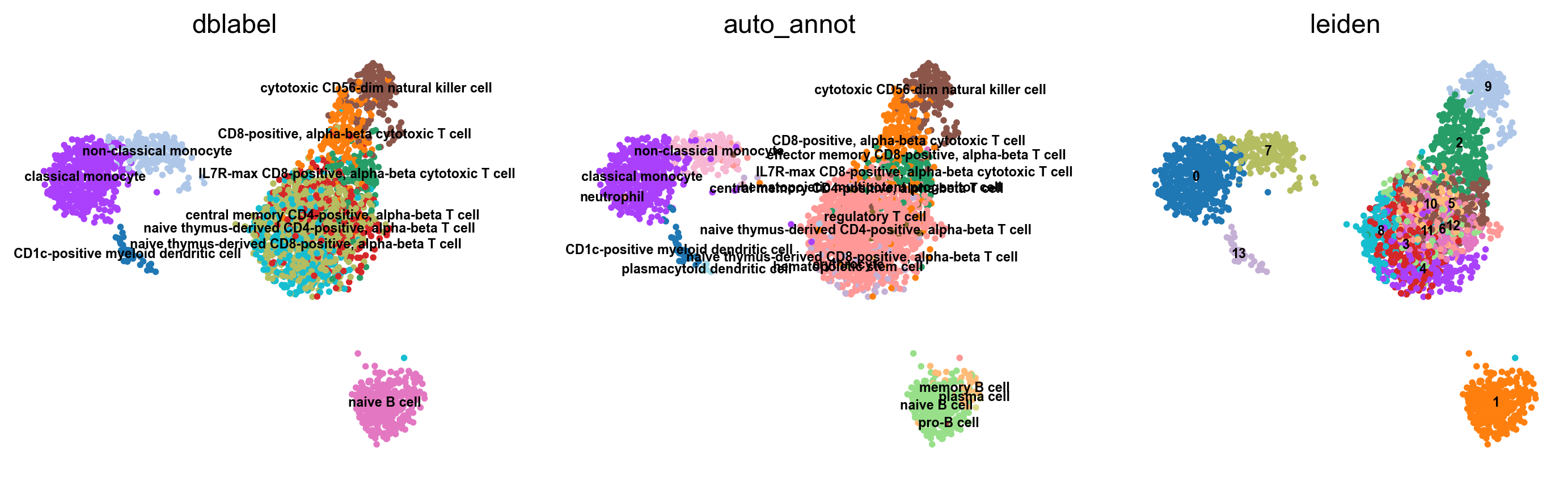
WARNING: saving figure to file figures/umap.auto_annot_tutorial_svm_dblabel.png
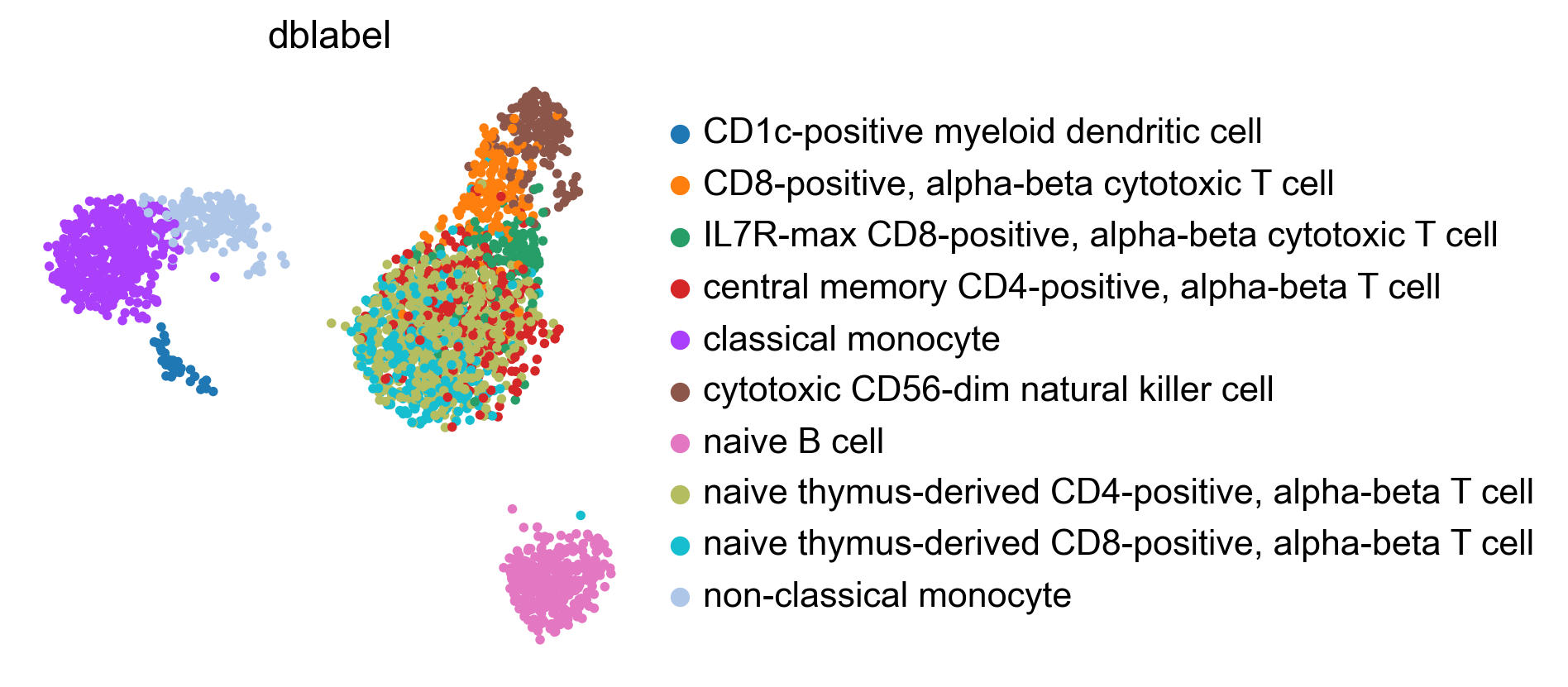
WARNING: saving figure to file figures/umap.auto_annot_tutorial_svm_auto_annot.png
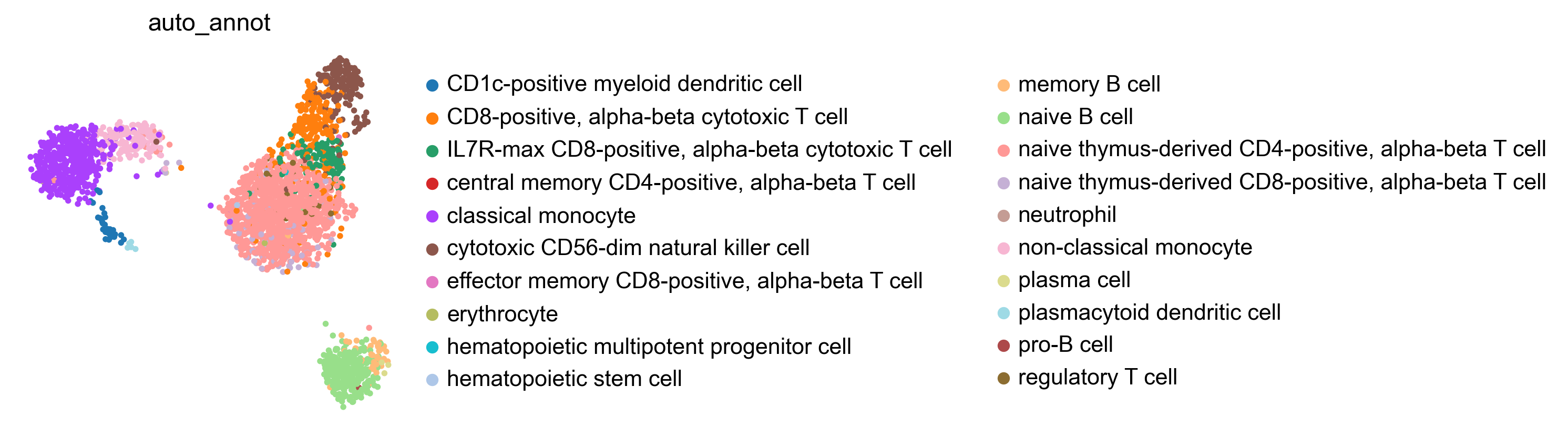
WARNING: saving figure to file figures/umap.auto_annot_tutorial_svm_leiden.png
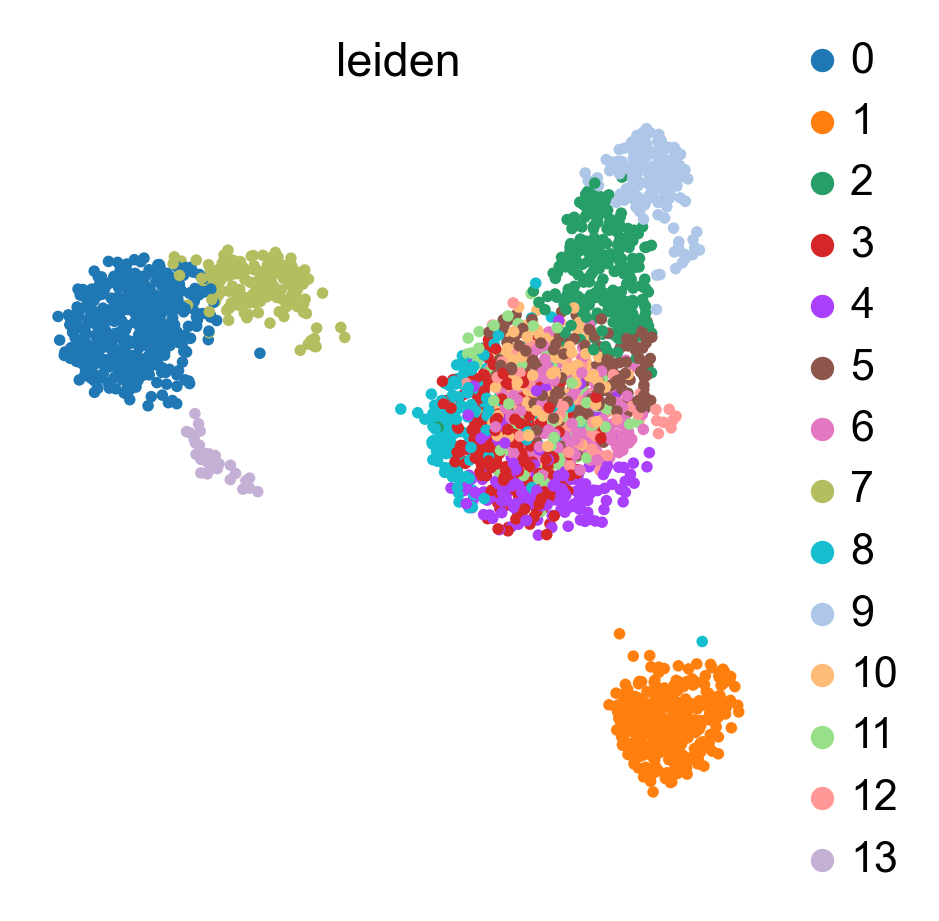
Data type cannot be displayed: application/vnd.plotly.v1+json

Let’s try with a specified gene set¶
Gene sets from GeMS or user specified genesets, can, if carefully chosen, significantly decrease computation time, withouth leading to a performance decrease.
[30]:
analysis_name = "auto_annot_tutorial_geneset"
celltype = "dblabel" # column name of celltype of interest
method = "linear" # rbf or linear or sgd, rbf extremely slow cannot recommend, logistic_regression recommended, as you can get probabilities, random_forest is a fast but not very powerful option (in current implementation)
merge = "scanorama"
use_raw = False
[31]:
annotSigns = bc.datasets.load_immune_signatures()
allGenes = lambda annotSigns: [item for sublist in annotSigns for item in sublist]
genes_to_use = list(set(allGenes(list(annotSigns.values()))))
display(genes_to_use[:10])
['MLANA',
'LEF1',
'MMRN1',
'GZMA',
'MCM2',
'FGFBP2',
'KRT18',
'PCNA',
'TRDC',
'FABP4']
[32]:
adata_test = adata_test_orig.copy()
[33]:
adata_train, adata_test_corrected = bc.tl.auto_annot.merge_data(
adata_train_list, adata_test, genes_to_use=genes_to_use, merge=merge
)
merging with scanorama
using scanorama rn
Found 77 genes among all datasets
[[0. 0.69 0.52]
[0. 0. 0.98]
[0. 0. 0. ]]
Processing datasets (1, 2)
Processing datasets (0, 1)
Processing datasets (0, 2)
integrating training set
[34]:
classifier, scaler = bc.tl.auto_annot.fit(adata_train, method, celltype)
[35]:
adata_predicted = bc.tl.auto_annot.adata_predict(
classifier=classifier,
scaler=scaler,
adata_pred=adata_test_corrected,
adata_orig=adata_test_orig,
)
[36]:
adata_predicted.write("/tmp/adata_predicted_svm.h5ad")
[37]:
%matplotlib inline
bc.tl.auto_annot.report(adata_predicted, celltype, method, analysis_name,
False, merge, use_raw, genes_to_use, clustering = 'leiden')
besca.tl.auto_annot.report(...) is deprecated( besca > 2.3); please use besca.tl.report(...)
WARNING: saving figure to file figures/umap.ondata_auto_annot_tutorial_geneset.png
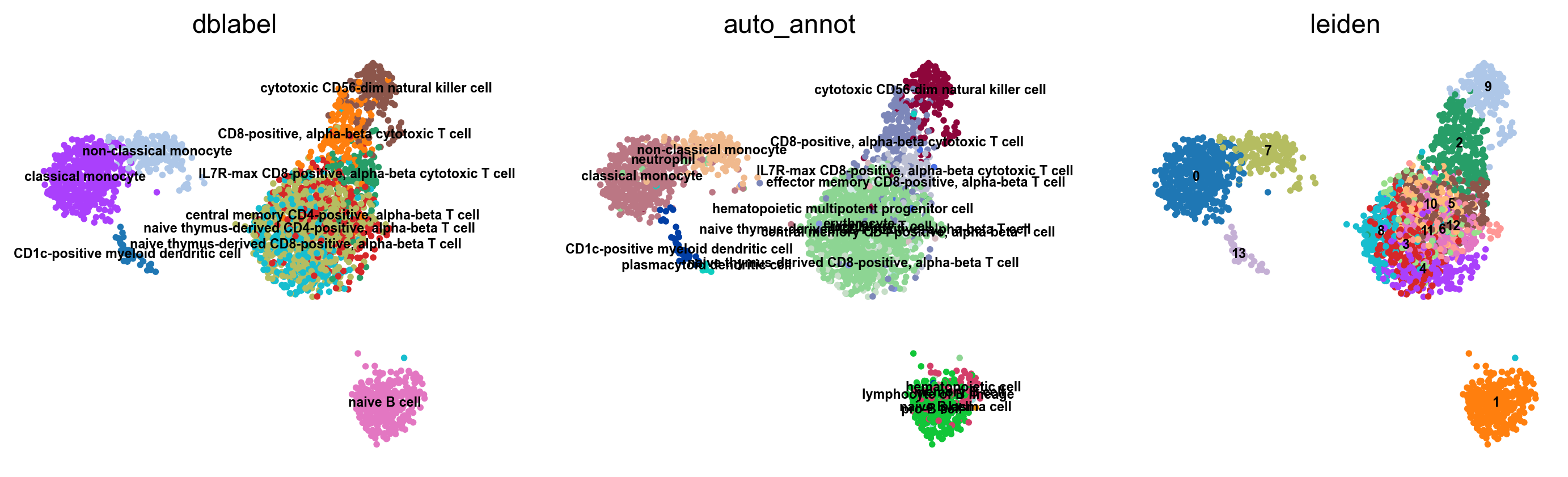
WARNING: saving figure to file figures/umap.auto_annot_tutorial_geneset_dblabel.png
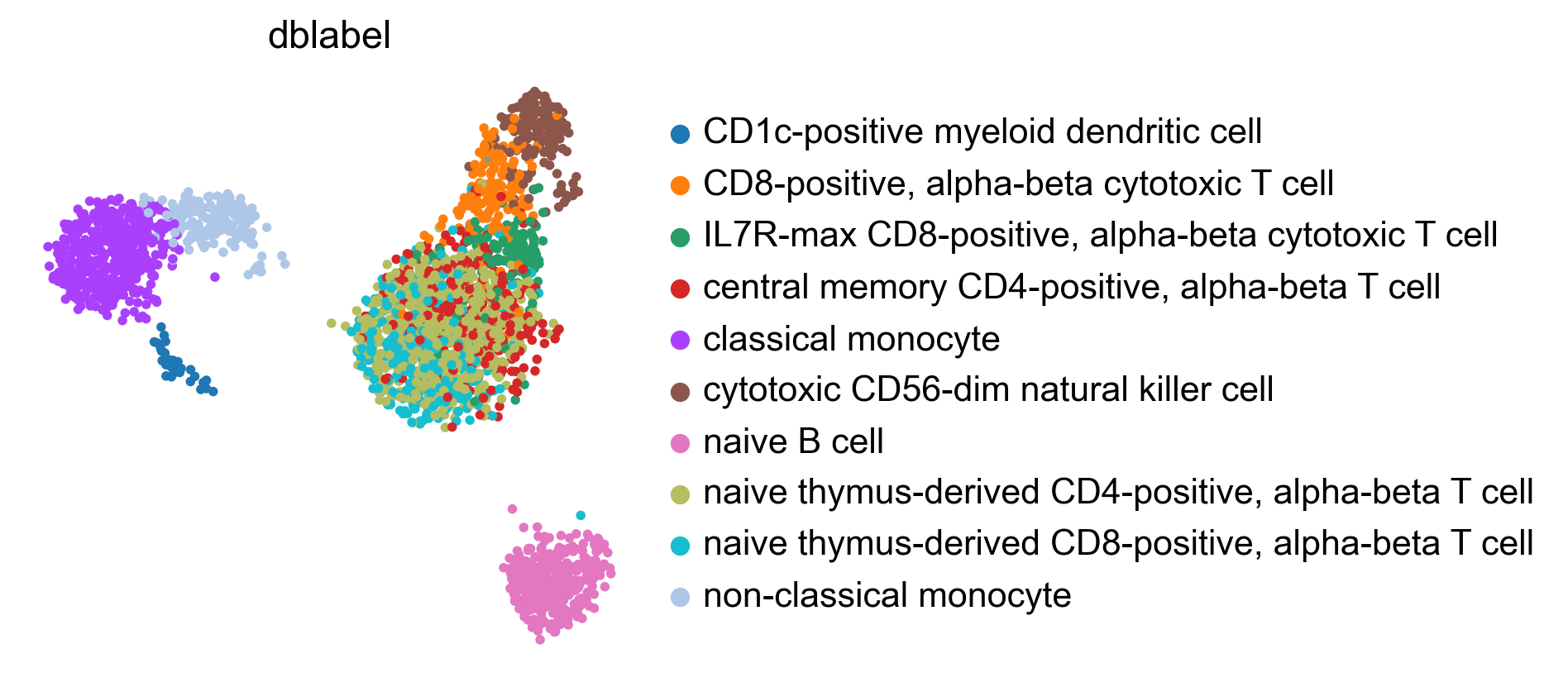
WARNING: saving figure to file figures/umap.auto_annot_tutorial_geneset_auto_annot.png
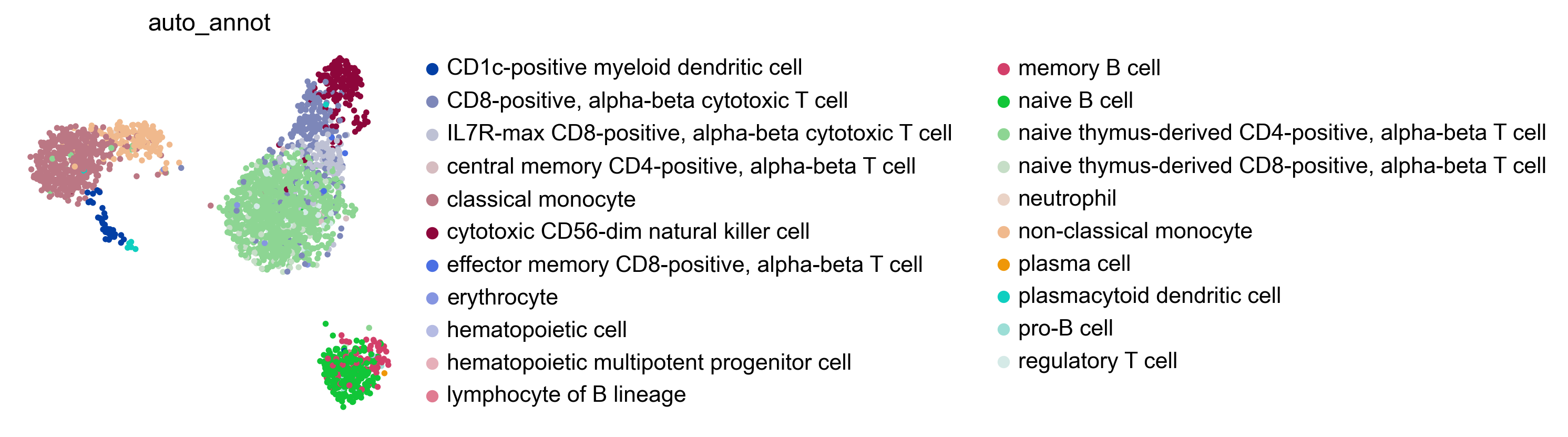
WARNING: saving figure to file figures/umap.auto_annot_tutorial_geneset_leiden.png
Data type cannot be displayed: application/vnd.plotly.v1+json