library(cd8ippred)
library(Biobase)
#> Loading required package: BiocGenerics
#>
#> Attaching package: 'BiocGenerics'
#> The following objects are masked from 'package:stats':
#>
#> IQR, mad, sd, var, xtabs
#> The following objects are masked from 'package:base':
#>
#> anyDuplicated, aperm, append, as.data.frame, basename, cbind,
#> colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find,
#> get, grep, grepl, intersect, is.unsorted, lapply, Map, mapply,
#> match, mget, order, paste, pmax, pmax.int, pmin, pmin.int,
#> Position, rank, rbind, Reduce, rownames, sapply, setdiff, sort,
#> table, tapply, union, unique, unsplit, which.max, which.min
#> Welcome to Bioconductor
#>
#> Vignettes contain introductory material; view with
#> 'browseVignettes()'. To cite Bioconductor, see
#> 'citation("Biobase")', and for packages 'citation("pkgname")'.
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following object is masked from 'package:Biobase':
#>
#> combine
#> The following objects are masked from 'package:BiocGenerics':
#>
#> combine, intersect, setdiff, union
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(tidyr)
library(ggplot2)This is an example of how to apply CD8 immune phenotype model to data.
Warning: Only apply this to RNA-Seq data processed with Roche-internal RNA-Seq pipeline (e.g., processed EDIS data). When in doubt, compare your data distribution to the example data.
We are using simulated data sampled from the training set. First we need Variance-stabilizing transformation.
cd8ip_simulated_counts <- load_simulated_counts()
vst_m <- compute_vst(exprs(cd8ip_simulated_counts))
#> Warning: replacing previous import 'S4Arrays::makeNindexFromArrayViewport' by
#> 'DelayedArray::makeNindexFromArrayViewport' when loading 'SummarizedExperiment'
#> converting counts to integer modeCreate a data.frame.
# set rownames to symbol names where available;
# the model uses only a subset of genes with symbols available.
rownames(vst_m) <- ifelse(
is.na(fData(cd8ip_simulated_counts)$symbol) | fData(cd8ip_simulated_counts)$symbol == "",
rownames(vst_m),
fData(cd8ip_simulated_counts)$symbol
)
# make rownames unqiue; the model uses only a subset of genes
# without symbol duplication issues
rownames(vst_m) <- make.names(rownames(vst_m), unique = TRUE)
# transform to a data.frame()
vst_df <- as.data.frame(t(vst_m))
# data preview
vst_df[1:6, 1:6]
#> TSPAN6 TNMD DPM1 SCYL3 C1orf112 FGR
#> ffc0 10.63564 5.578913 10.95236 9.547618 9.847032 9.118320
#> fd98 11.15683 5.691649 11.51214 9.101428 10.451604 7.894474
#> 46d7 9.26342 5.927867 11.25951 9.320227 11.390358 8.041969
#> c877 9.42878 5.283350 10.94896 9.794054 10.569744 7.353412
#> eddf 11.56120 5.283350 11.14481 9.083383 9.989589 7.390809
#> 3dba 10.66487 6.326243 10.20469 8.059314 10.308865 8.394831Predict immune phenotypes
pred <- predict_cd8ip(vst_df)
pred$truth <- cd8ip_simulated_counts$CD8IMMPH |>
tolower() |>
# we convert truth to a factor for yardstick ROC computations
factor()Prediction quality
# values above 6 should be interpreted with caution
summary(pred$n_out_of_range)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 0.0000 0.0000 0.0000 0.0375 0.0000 1.0000
# low-confidence predictions are set to NA
summary(is.na(pred$predicted.class))
#> Mode FALSE
#> logical 80Confusion matrix
with(
pred,
table(truth, predicted.class)
)
#> predicted.class
#> truth desert excluded inflamed
#> desert 15 5 0
#> excluded 1 18 1
#> inflamed 0 2 18View data.frame with predictions
head(pred)
#> prob.inflamed prob.excluded prob.desert predicted.class n_out_of_range
#> ffc0 0.10191362 0.1731693 0.7249171 desert 0
#> fd98 0.21714803 0.5062805 0.2765715 excluded 0
#> 46d7 0.19156818 0.5225662 0.2858657 excluded 0
#> c877 0.15159548 0.2483887 0.6000158 desert 0
#> eddf 0.10275997 0.3010333 0.5962067 desert 0
#> 3dba 0.05888132 0.2506064 0.6905123 desert 0
#> truth
#> ffc0 desert
#> fd98 desert
#> 46d7 desert
#> c877 desert
#> eddf desert
#> 3dba desertAUC scores
Please note, this is not real data. These AUC scores are only an example.
auc_df <- pred |>
filter(!is.na(truth)) |>
pivot_longer(starts_with("prob."), names_prefix = "prob.", names_to = ".level", values_to = "prob") |>
mutate(positive = as.factor(truth == .level)) |>
group_by(.level) |>
yardstick::roc_auc(positive, prob, event_level = "second") |>
rename(AUC = .estimate, class = .level) |>
select(class, AUC) %>%
mutate(AUC_text = paste0("AUC=", round(AUC, 3)))
select(auc_df, -AUC_text)
#> # A tibble: 3 × 2
#> class AUC
#> <chr> <dbl>
#> 1 desert 0.945
#> 2 excluded 0.854
#> 3 inflamed 0.996ROC curve
pheno_col <- c(
inflamed = "#d95f02",
excluded = "#1b9e77",
desert = "#7570b3"
)
pred |>
filter(!is.na(truth)) |>
# column names should match levels of the truth factor
yardstick::roc_curve(truth, all_of(paste0("prob.", levels(pred$truth)))) |>
mutate(class = .level) |>
ggplot() +
aes(x = 1 - specificity, y = sensitivity, col = class) +
geom_path() +
geom_abline(lty = 3) +
coord_equal() +
geom_text(aes(label = AUC_text), x = 0.55, y = 0.1, data = auc_df, show.legend = FALSE) +
facet_wrap(~class, ncol = 2) +
scale_color_manual(values = pheno_col) +
theme_bw()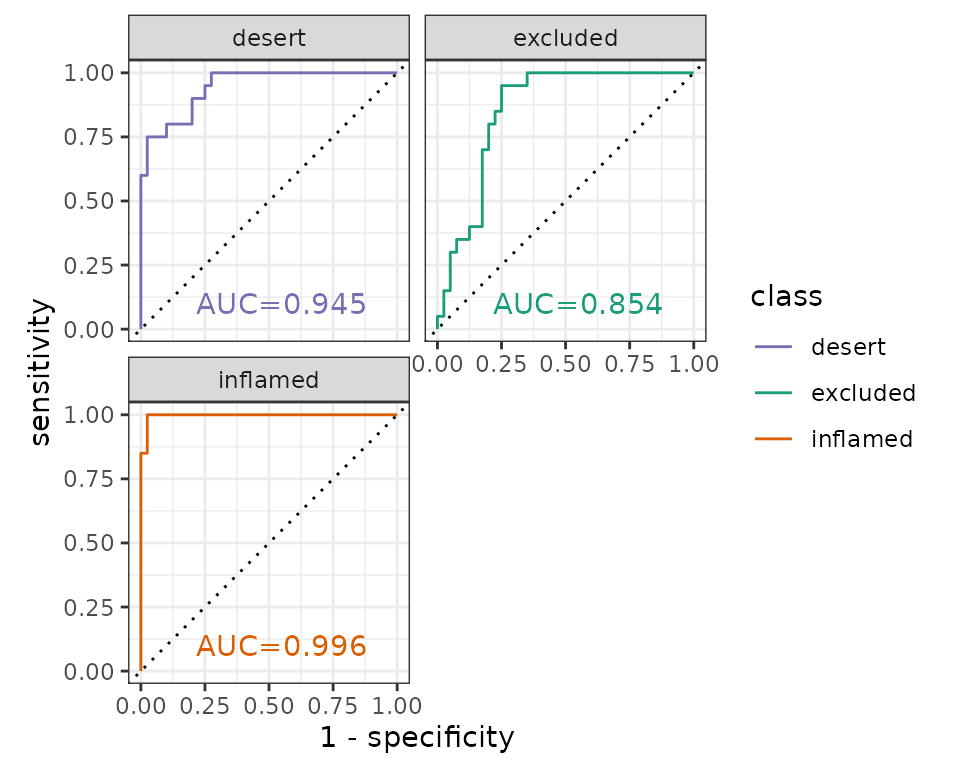
Session info
devtools::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.3.1 (2023-06-16)
#> os Ubuntu 22.04.3 LTS
#> system x86_64, linux-gnu
#> ui X11
#> language en
#> collate C.UTF-8
#> ctype C.UTF-8
#> tz UTC
#> date 2023-09-05
#> pandoc 2.19.2 @ /usr/bin/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> ! package * version date (UTC) lib source
#> P abind 1.4-5 2016-07-21 [?] RSPM (R 4.3.0)
#> P assertthat 0.2.1 2019-03-21 [?] RSPM (R 4.3.0)
#> P Biobase * 2.60.0 2023-04-25 [?] Bioconductor
#> P BiocGenerics * 0.46.0 2023-04-25 [?] Bioconductor
#> P BiocManager 1.30.22 2023-08-08 [?] RSPM (R 4.3.0)
#> P BiocParallel 1.34.2 2023-05-22 [?] Bioconductor
#> P bitops 1.0-7 2021-04-24 [?] RSPM (R 4.3.0)
#> P bslib 0.5.1 2023-08-11 [?] RSPM (R 4.3.0)
#> P cachem 1.0.8 2023-05-01 [?] RSPM (R 4.3.0)
#> P callr 3.7.3 2022-11-02 [?] RSPM (R 4.3.0)
#> cd8ippred * 0.1.0 2023-09-05 [1] local
#> P cli 3.6.1 2023-03-23 [?] RSPM (R 4.3.0)
#> P codetools 0.2-19 2023-02-01 [?] CRAN (R 4.3.1)
#> P colorspace 2.1-0 2023-01-23 [?] RSPM (R 4.3.0)
#> P crayon 1.5.2 2022-09-29 [?] RSPM (R 4.3.0)
#> P DelayedArray 0.26.7 2023-07-28 [?] Bioconductor
#> P desc 1.4.2 2022-09-08 [?] RSPM (R 4.3.0)
#> P DESeq2 1.40.2 2023-06-23 [?] Bioconductor
#> P devtools 2.4.5 2022-10-11 [?] RSPM (R 4.3.0)
#> P digest 0.6.33 2023-07-07 [?] RSPM (R 4.3.0)
#> P dplyr * 1.1.2 2023-04-20 [?] RSPM (R 4.3.0)
#> P ellipsis 0.3.2 2021-04-29 [?] RSPM (R 4.3.0)
#> P evaluate 0.21 2023-05-05 [?] RSPM (R 4.3.0)
#> P fansi 1.0.4 2023-01-22 [?] RSPM (R 4.3.0)
#> P farver 2.1.1 2022-07-06 [?] RSPM (R 4.3.0)
#> P fastmap 1.1.1 2023-02-24 [?] RSPM (R 4.3.0)
#> P foreach 1.5.2 2022-02-02 [?] RSPM (R 4.3.0)
#> P fs 1.6.3 2023-07-20 [?] RSPM (R 4.3.0)
#> P generics 0.1.3 2022-07-05 [?] RSPM (R 4.3.0)
#> P GenomeInfoDb 1.36.2 2023-08-25 [?] Bioconductor
#> P GenomeInfoDbData 1.2.10 2023-08-31 [?] Bioconductor
#> P GenomicRanges 1.52.0 2023-04-25 [?] Bioconductor
#> P ggplot2 * 3.4.3 2023-08-14 [?] RSPM (R 4.3.0)
#> P glmnet 4.1-8 2023-08-22 [?] RSPM (R 4.3.0)
#> P glue 1.6.2 2022-02-24 [?] RSPM (R 4.3.0)
#> P gtable 0.3.4 2023-08-21 [?] RSPM (R 4.3.0)
#> P highr 0.10 2022-12-22 [?] RSPM (R 4.3.0)
#> P htmltools 0.5.6 2023-08-10 [?] RSPM (R 4.3.0)
#> P htmlwidgets 1.6.2 2023-03-17 [?] RSPM (R 4.3.0)
#> P httpuv 1.6.11 2023-05-11 [?] RSPM (R 4.3.0)
#> P IRanges 2.34.1 2023-06-22 [?] Bioconductor
#> P iterators 1.0.14 2022-02-05 [?] RSPM (R 4.3.0)
#> P jquerylib 0.1.4 2021-04-26 [?] RSPM (R 4.3.0)
#> P jsonlite 1.8.7 2023-06-29 [?] RSPM (R 4.3.0)
#> P knitr 1.43 2023-05-25 [?] RSPM (R 4.3.0)
#> P labeling 0.4.3 2023-08-29 [?] RSPM (R 4.3.0)
#> P later 1.3.1 2023-05-02 [?] RSPM (R 4.3.0)
#> P lattice 0.21-8 2023-04-05 [?] CRAN (R 4.3.1)
#> P lifecycle 1.0.3 2022-10-07 [?] RSPM (R 4.3.0)
#> P locfit 1.5-9.8 2023-06-11 [?] RSPM (R 4.3.0)
#> P magrittr 2.0.3 2022-03-30 [?] RSPM (R 4.3.0)
#> P Matrix 1.6-1 2023-08-14 [?] RSPM (R 4.3.0)
#> P MatrixGenerics 1.12.3 2023-07-30 [?] Bioconductor
#> P matrixStats 1.0.0 2023-06-02 [?] RSPM (R 4.3.0)
#> P memoise 2.0.1 2021-11-26 [?] RSPM (R 4.3.0)
#> P mime 0.12 2021-09-28 [?] RSPM (R 4.3.0)
#> P miniUI 0.1.1.1 2018-05-18 [?] RSPM (R 4.3.0)
#> P munsell 0.5.0 2018-06-12 [?] RSPM (R 4.3.0)
#> P pillar 1.9.0 2023-03-22 [?] RSPM (R 4.3.0)
#> P pkgbuild 1.4.2 2023-06-26 [?] RSPM (R 4.3.0)
#> P pkgconfig 2.0.3 2019-09-22 [?] RSPM (R 4.3.0)
#> P pkgdown 2.0.7 2022-12-14 [?] RSPM (R 4.3.0)
#> P pkgload 1.3.2.1 2023-07-08 [?] RSPM (R 4.3.0)
#> P prettyunits 1.1.1 2020-01-24 [?] RSPM (R 4.3.0)
#> P processx 3.8.2 2023-06-30 [?] RSPM (R 4.3.0)
#> P profvis 0.3.8 2023-05-02 [?] RSPM (R 4.3.0)
#> P promises 1.2.1 2023-08-10 [?] RSPM (R 4.3.0)
#> P ps 1.7.5 2023-04-18 [?] RSPM (R 4.3.0)
#> P purrr 1.0.2 2023-08-10 [?] RSPM (R 4.3.0)
#> P R6 2.5.1 2021-08-19 [?] RSPM (R 4.3.0)
#> P ragg 1.2.5 2023-01-12 [?] RSPM (R 4.3.0)
#> P Rcpp 1.0.11 2023-07-06 [?] RSPM (R 4.3.0)
#> P RCurl 1.98-1.12 2023-03-27 [?] RSPM (R 4.3.0)
#> P remotes 2.4.2.1 2023-07-18 [?] RSPM (R 4.3.0)
#> renv 1.0.2 2023-08-15 [2] RSPM (R 4.3.0)
#> P rlang 1.1.1 2023-04-28 [?] RSPM (R 4.3.0)
#> P rmarkdown 2.24 2023-08-14 [?] RSPM (R 4.3.0)
#> P rprojroot 2.0.3 2022-04-02 [?] RSPM (R 4.3.0)
#> P S4Arrays 1.0.5 2023-07-24 [?] Bioconductor
#> P S4Vectors 0.38.1 2023-05-02 [?] Bioconductor
#> P sass 0.4.7 2023-07-15 [?] RSPM (R 4.3.0)
#> P scales 1.2.1 2022-08-20 [?] RSPM (R 4.3.0)
#> P sessioninfo 1.2.2 2021-12-06 [?] RSPM (R 4.3.0)
#> P shape 1.4.6 2021-05-19 [?] RSPM (R 4.3.0)
#> P shiny 1.7.5 2023-08-12 [?] RSPM (R 4.3.0)
#> P stringi 1.7.12 2023-01-11 [?] RSPM (R 4.3.0)
#> P stringr 1.5.0 2022-12-02 [?] RSPM (R 4.3.0)
#> P SummarizedExperiment 1.30.2 2023-06-06 [?] Bioconductor
#> P survival 3.5-7 2023-08-14 [?] RSPM (R 4.3.0)
#> P systemfonts 1.0.4 2022-02-11 [?] RSPM (R 4.3.0)
#> P textshaping 0.3.6 2021-10-13 [?] RSPM (R 4.3.0)
#> P tibble 3.2.1 2023-03-20 [?] RSPM (R 4.3.0)
#> P tidyr * 1.3.0 2023-01-24 [?] RSPM (R 4.3.0)
#> P tidyselect 1.2.0 2022-10-10 [?] RSPM (R 4.3.0)
#> P urlchecker 1.0.1 2021-11-30 [?] RSPM (R 4.3.0)
#> P usethis 2.2.2 2023-07-06 [?] RSPM (R 4.3.0)
#> P utf8 1.2.3 2023-01-31 [?] RSPM (R 4.3.0)
#> P vctrs 0.6.3 2023-06-14 [?] RSPM (R 4.3.0)
#> P withr 2.5.0 2022-03-03 [?] RSPM (R 4.3.0)
#> P xfun 0.40 2023-08-09 [?] RSPM (R 4.3.0)
#> P xtable 1.8-4 2019-04-21 [?] RSPM (R 4.3.0)
#> P XVector 0.40.0 2023-04-25 [?] Bioconductor
#> P yaml 2.3.7 2023-01-23 [?] RSPM (R 4.3.0)
#> P yardstick 1.2.0 2023-04-21 [?] RSPM (R 4.3.0)
#> P zlibbioc 1.46.0 2023-04-25 [?] Bioconductor
#>
#> [1] /tmp/Rtmp4vwKTI/temp_libpath143d3eff14e6
#> [2] /home/runner/.cache/R/renv/library/cd8ippred-05538736/R-4.3/x86_64-pc-linux-gnu
#> [3] /home/runner/.cache/R/renv/sandbox/R-4.3/x86_64-pc-linux-gnu/5cd49154
#>
#> P ── Loaded and on-disk path mismatch.
#>
#> ──────────────────────────────────────────────────────────────────────────────